Introduction
Drug Target Commons (DTC) is a community based effort to improve the consensus on drug-target interactions. Our system contains two types of bioactivity data i.e. annotated and annotated. Initial search on 'Home page' allows the users to view existing (unannotated) bioactivity data using compound, target or publications. However, users may submit suggestions to edit/add bioactivity data and MicroBAO annotations.
User Information/login
Search
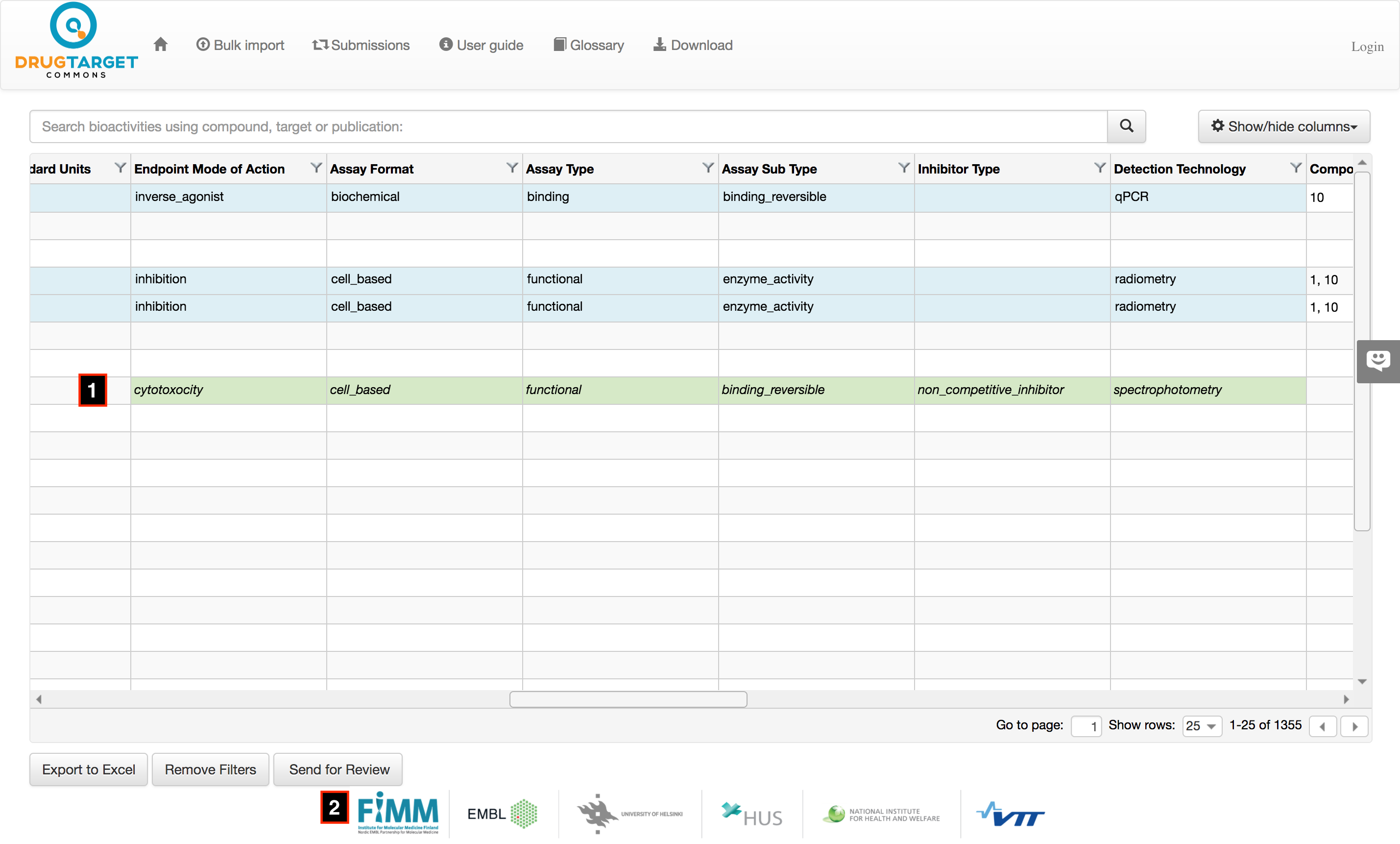
Search keyword are matched with annotated/unannotated compounds, targets and publications, therefor you get a combined set of results from which you select the preferred result.
Compound search
Compound search can be by name or synonyms, standard inchikey or any of 26 IDs (Chembl, bindingdb, molport, surechembl, actor, nmrshiftdb2, chebi, hmdb, fdasrs, selleck, lincs, drugbank, nih_ncc, pharmgkb, ibm, nikkaji, pubchem, emolecules, zinc, recon, mcule, pubchem_dotf, gtopdb, pdb, pubchem_tpharma, kegg_ligand). Partial and exact searches are supportable for compound name or synonyms; while inchikey or IDs must have exact match. After entering compound name or ID, user must have to click on search button (or press enter key) to search it in present database.
Target search
Target search can be made by target name, gene name and ID (UniProtKB-AC, UniProtKB-ID, EMBL-CDS, Ensembl, PIR, PubMed, NCBI-taxon, Ensembl_TRS, MIM, UniGene, UniRef50, RefSeq, GO, PDB, UniParc, Additional PubMed, EMBL, GI, GeneID, Chembl, UniRef100, UniRef90, Ensembl_PRO) . Rest of specifications are more or less similar to compound search. Again same filter on target types. User may perform filtering, searcing, reordering of columns and export to excell to the resulting records.
Publication search
Publication search can be done using "Pubmed ID". If "pubmed ID" information doesn’t exists in our present database user may manually enter all the fields. If ‘pubmed ID’ information exists in our system, than some of the information about compound, targets, bioactivities, assays and publication will be retrieved automatically. However MicroBAO and few more fields will be left blank for user. Again, User may filter out irrelevant compounds or targets.
Add annotations
Annotations can be added by uploading a excel file(suitable for large data) or directly from search results
-
Click on the “Download Template” clink and down the excel file template.
-
Fill the columns with required annotated data
-
Click ' Send for Review '
Uploaded Annotations can be previewed and edited before submitting
Add Annotations directly from search results